Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
TaubCONNECT Research Perspectives:
March 2017
#2 Polygenic Risk Scores in Familial Alzheimer Disease
 |  | |
| Asa Abeliovich, MD, PhD | Herve Rhinn, PhD |
Genome-wide association studies (GWAS) have uncovered genetic determinants for common neurodegenerative disorders, such as Alzheimer’s disease (AD), and thus provided important and tractable insights into potential mechanisms of pathogenesis. But less is known about mechanisms that underlie normal human brain aging, which is by far the most critical risk factor for such diseases. Drs. Asa Abeliovich and Herve Rhinn of the Taub Institute sought to apply some of the integrative genomics tools previously used for the analysis of disease, such as GWAS, to pursue determinants and mechanisms of aging in human cortex, as published in Cell Systems this month.
Aging phenomena, such as skin wrinkling or cognitive changes, appear surprisingly variable across the human population, and the researchers hypothesized that some such heterogeneity may be due to underlying genetic diversity; in other words, that some common human genetic variants may lead to an individual appearing older or younger than predicted, relative to his or her cohort, at any given chronological age. To pursue genetic determinants of aging in the human cortex, the authors mined large existing human genomics and transcriptomics datasets, to identify all of the transcripts that are predictive of age (that correlate with aging) in the human frontal cortex. Subsequently, for each individual in a cohort, a score was obtained - termed Δ-aging – that quantified whether an individual appeared "older" or "younger" than predicted for their chronological age, relative to the rest of their cohort (based on an analysis of the set of age-dependent brain transcripts). Applying this approach to a large-scale collection of autopsied human frontal cortex tissue samples, a GWAS was performed, and identified the TMEM106B gene locus as by far the most significant regulator of human frontal cortex aging. Carriers of risk variants at the TMEM106B gene locus can appear ~12 years older than carriers of protective variants.
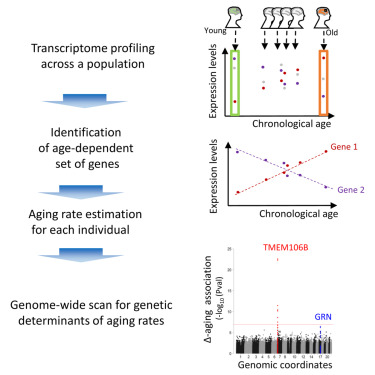
Graphical Abstract
The role of TMEM106B variants on brain aging appears highly selective, both anatomically and in terms of life stage: the effect is seen selectively in frontal cortex but not other brain regions, and primarily during late-life aging (beyond 65 years old). Strikingly, TMEM106B variants that lead to accelerated aging in this analysis of the general human population had previously been associated with an increased risk for frontotemporal dementia, a rare neurodegenerative disease of frontal cortex aging. It is possible that the effect of TMEM106B variants on FTD risk is largely driven by accelerated aging of the frontal cortex, as the disease is highly age-dependent. Finally, a detailed analysis of the transcriptome changes in frontal cortex tissue from individuals who carry the risk TMEM106B genetic variants, associated with accelerated aging, suggests an important role for altered inflammation, a process termed “inflammaging”.
This study by Drs. Rhinn and Abeliovich was also covered in CUMC Newsroom this month.
Asa Abeliovich, MD, PhD
Associate Professor of Pathology and Cell Biology, and Neurology (in the Taub Institute)
aa900@cumc.columbia.edu
Herve Rhinn, PhD
Assistant Professor of Pathology and Cell Biology
hr2239@cumc.columbia.edu
Polygenic Risk Scores in Familial Alzheimer Disease
 |  |
| Giuseppe Tosto, MD, PhD | Richard Mayeux, MD, MSc |
Over the past decade, researchers have identified several loci associated with late-onset Alzheimer's disease (LOAD) through genome-wide association studies (GWAS) and sequencing studies, as detailed in a previous review by Drs. Giuseppe Tosto and Christiane Reitz. Over 20 susceptibility loci have been replicated or newly identified by a large GWAS performed by the International Genomics of Alzheimer's Project (IGAP), yet the effect sizes associated with all these new variants have been small, suggesting that a large part of the genetic component of LOAD remains unexplained.
Polygenic risk scores (genetic risk scores [GRS]) represent an alternative strategy to summarize sparse genetic information and identify genetic risk profiles. Previous investigations imply that sporadic LOAD is characterized by a significant polygenic component, yet the GRS has never been investigated in familial LOAD or in other ethnic groups with familial LOAD. Now published online in Neurology, Dr. Tosto and colleagues from the Taub Institute and CUMC, along with other institutions, tested the significance of a GRS in two cohorts of families multiply affected by LOAD: the National Institute of Aging-Late Onset Alzheimer's Disease (NIA-LOAD) family study and Estudio Familiar d Influencia Genetic en Alzheimer (EFIGA) study.
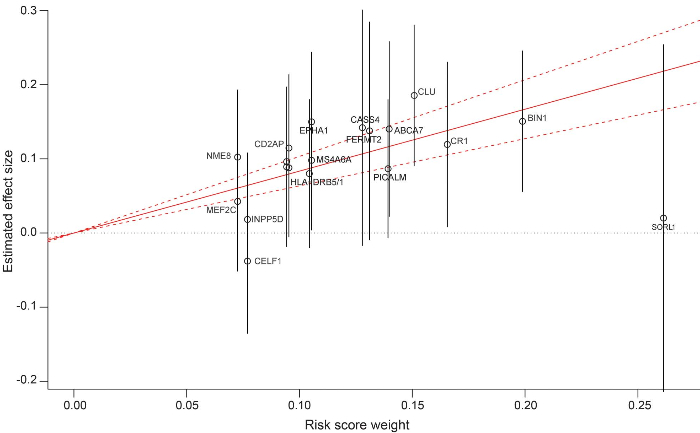
Figure 1. National Institute on Aging – Late-Onset Alzheimer ’ s Disease Family Study (NIA-LOAD) polygenic risk score (GRS) and its weighting coefficients. Construction of the NIA-LOAD polygenic risk score. Each single nucleotide polymorphism is plotted by the weighting coefficient extracted from the Inter- national Genomics of Alzheimer ’ s Project analysis (x axis) vs the estimated effect size (and 95% confidence interval [CI] depicted by vertical gray lines) obtained by the mixed model (y axis). The solid red line indicates the effect size estimate for the GRS on late-onset Alzheimer disease (dotted lines represent the 95% CI). SLC24A4 , ZCWPW1 , and PTK2B share the same position on the graph, and for graphic clarity are not labeled
Tosto et al. found that high GRS increased the risk of LOAD in families with multiple affected with the disease, and lowered the age-at-onset. The results were independent of the risk associated with the APOE e4 allele and regardless of the ethnic group. Further investigations will be required when high throughput sequence data became available for these large cohorts. Identifying individuals with high risk of developing the disease based on their genetic profile would benefit clinical trial enrollment as well as precision medicine for personalized drug development.
Giuseppe Tosto, MD, PhD
Postdoctoral Research Scientist (in the Taub Institute and the Gertrude H. Sergievsky Center)
gt2260@cumc.columbia.edu
Richard Mayeux, MD, MSc
Gertrude H. Sergievsky Professor of Neurology, Psychiatry and Epidemiology (in the Gertrude H. Sergievsky Center and in the Taub Institute)
rpm2@cumc.columbia.edu