Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
TaubCONNECT Research Perspectives:
Best Poster Presentations
Taub Institute Retreat November 2017
A CSF Proteomic Screen Links Retromer to Alzheimer's Pathogenic Pathways and Suggests Endosomal-Trafficking Biomarkers
Jessi Neufeld1, Emily Chen2, Larry Honig1,3,4, Sabrina Simoes1, Scott Small1,5.
1 Taub Institute, College of Physicians and Surgeons, Columbia University;
2 Department of Pharmacology, Columbia University Medical Center;
3 Department of Neurology, Columbia University Medical Center;
4 Gertrude H. Sergievsky Center, Columbia University Medical Center;
5 New York State Psychiatric Institute;

Jessi Neufeld in front of her winning poster at the 8th Annual Taub Institute Retreat
While biomarkers exist for the histological hallmarks of Alzheimer's disease (AD)—amyloid plaques and neurofibrillary tangles—currently lacking are biomarkers for endosomal traffic jams, a cytopathological hallmark now thought to play a pathogenic role in AD. With this goal in mind, we used the CAMK2A-CRE system to genetically engineer knockouts of VPS35, retromer’s core protein, in the forebrain neurons of mice. We isolated high quality CSF from KOs and controls, performed a proteomic screen using mass spectrometry, and subsequently validated hits via immunoblot in additional cohorts. Methods: CSF was collected from 6-month-old male mice, assessed for blood contamination via hemoglobin ELISA, and pooled into biological replicates of 30uL each (5 KO vs 4 control). Pooled CSF samples were analyzed by shotgun mass spectrometry approach using the Thermo Orbitrap Fusion Tribrid mass spectrometer. Results: In total, 1505 proteins were identified and over 30 proteins were significantly altered in the CSF of the KO mice. Two classes of elevated proteins were of particular interest: the first is a group of apolipoproteins (ApoE, ApoJ/clusterin, and ApoD), which have established roles in cholesterol transport and have previously been implicated in AD pathology. The second group comprises N-terminal fragments of substrates (CHL1, APLP1, and APLP2) of BACE1, the enzyme which initiates amyloidogenic processing of APP in the endosome. To confirm and validate these findings, we then turned to immunoblot analysis using CSF isolated from a separate cohort of mice. Consistent with the mass spectrometry data, we observed significant increases in CSF levels of ApoE, clusterin, and CHL1 and a trend toward increase in APLP1. Discussion: Our results suggest that endosomal trafficking and cholesterol metabolism, two genetic factors linked to AD, might interact in the brain. We are currently testing our hits in human CSF samples to determine whether they can be developed as biomarkers of endosomal-trafficking.
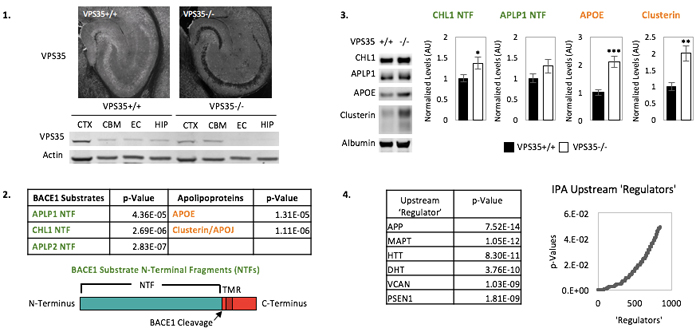 Figure: BACE1 substrate N-terminal fragments (NTFs) and apolipoproteins are elevated in the CSF of mice lacking neuronal VPS35. 1. VPS35 is markedly reduced in the hippocampi of VPS35fl/fl Camkα-cre KO mice. 2. Among over 30 hits, two families of proteins linked to AD pathology are elevated in the CSF of VPS35 KO mice: BACE1 substrate N-terminal fragments (NTFs) and apolipoproteins. 3. NTFs of BACE1 Substrates CHL1 and APLP1 and apolipoproteins APOE and clusterin are validated in CSF from a new cohort of mice. 4. Ingenuity Pathway Analysis (IPA) reveals AD pathogenic proteins among top 'upstream regulators' of CSF-based protein alterations in VPS35 KO mice.
|
Jessi Neufeld
Student, Graduate Program in Pathobiology and Molecular Medicine, Columbia University Medical Center
jn2325@cumc.columbia.edu
Microglia Identity in the Aged and AD Human Brain
Olah M1,2, Patrick E3, Villani AC2,4, Menon V1, Xu J2, White CC2, Ryan KJ, Piehowski P5, Kapasi A6, Nejad P2, Cimpean M, Connor S1,2, Yung CJ1,2,Frangieh M, McHenry A, Elyaman W1,2, Petyuk V5, Schneider JA6, Bennett DA6, De Jager PL1,2 and Bradshaw EM1,2.
1 Center for Translational & Computational Neuroimmunology, Department of Neurology, Columbia University Medical Center, New York City, NY
2 Program in Medical and Population Genetics, Broad Institute, 415 Main Street Cambridge, MA
3 School of Mathematics and Statistics, The Westmead Institute for Medical Research, The University of Sydney, Sydney, Australia
4 Massachusetts General Hospital, Boston, MA
5 Pacific Northwest National Laboratory, 902 Battelle Boulevard, Richland, WA
6 Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, Illinois, USA

Marta Olah, PhD
Microglia, the resident innate immune cells of the brain, have been implicated by genetic studies in the pathogenesis of age-related neurodegenerative disease. In order to further our understanding of microglial contribution to late onset neurodegenerative diseases, we established the first transcriptomic landscape of aged human microglia at the population, as well as at the single cell level. We have identified a set of genes that is preferentially expressed by microglia in the aged human brain; the expression of this transcriptomic signature was confirmed at the protein level. This gene set was found to be enriched in susceptibility genes for Alzheimer's disease (AD) and multiple sclerosis, to be increased with advancing age, and to be reduced by the AD protective APOEε2 haplotype. These findings confirm the existence of a unique microglia phenotype in the aged human brain and its involvement in the pathological processes associated with brain aging.
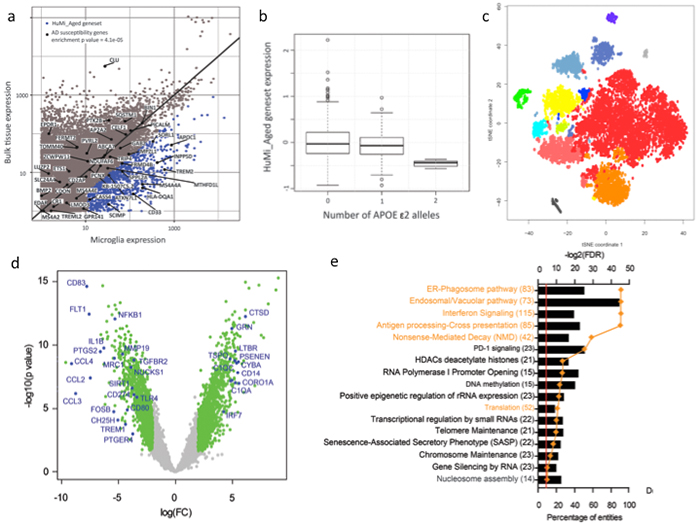 Figure: Microglia identity in the aged human brain. a) We have identified the genes that are preferentially expressed in microglia in the aged human brain (blue dots), and named them the HuMi_Aged geneset. This gene set was significantly enriched in AD susceptibility genes. b) The carriers of APOE ε2 alleles have significantly lower expression of the HuMi_Aged geneset. c) Single cell RNA-sequencing revealed the phenotypic heterogeneity of aged human microglia. d) Differential gene expression analysis between middle aged and aged human microglia identified the genes that were down and upregalated in microglia with aging. e) Gene ontology analysis of the upregulated genes identified the pathways that gain in function with microglia aging.
|
Marta Olah, PhD
Instructor in Neurology at the Columbia University Medical Center
mo2738@cumc.columbia.edu