Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
In the Lab:
Hans-Ulrich Klein, PhD

Hans-Ulrich Klein, PhD
The advent of new technologies and analytic approaches is beginning to provide an unbiased genome-wide look at epigenomic, transcriptomic and proteomic features of the aging human brain. Recently generated multi-omic Alzheimer’s disease (AD) datasets are of immense complexity comprising molecular signatures from different cell types and different brain regions from several hundred subjects at different stages of the disease. As a computational scientist, my research focuses on new analytic methods for integrating multi-omic data to enrich our understanding of AD pathogenesis and to possibly guide development of new therapeutic targets. Of particular interest, among others, are computational approaches to uncover how genetic risk variants exert their effects, methods for disentangling the roles of different cell types in disease processes, and structural causal models to infer dependencies among disease processes.
In a recent project together with Dr. Philip De Jager and the Center for Translational and Computational Neuroimmunology, we studied the epigenetic mark histone 3 lysine 9 acetylation (H3K9ac) in 669 aged human prefrontal cortices. We identified large-scale changes throughout the epigenome of the human AD brain and showed that tau-induced alterations of chromatin structure are much more profound than the changes that are attributable to amyloid pathology. The effect of these alterations propagated into the transcriptome. Our data supports the hypothesis that the nuclear lamina is a key element in mediating tau toxicity (Figure 1).
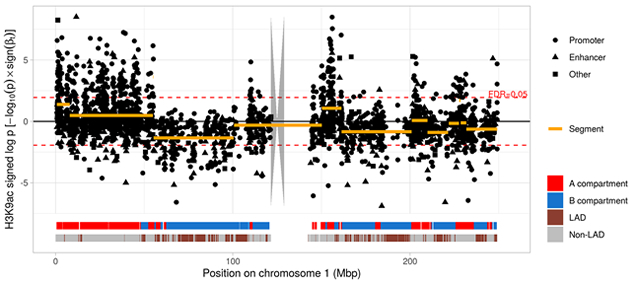 Figure 1. Manhattan plot from a histone acetylome-wide association study of tau pathology. Each dot represents a H3K9ac domain on chromosome 1. Log-transformed P values for tau are shown on the y-axis and were multiplied with the sign of the respective coefficient to distinguish between positive and negative associations. Large genomic segments (orange lines) covered multiple H3K9ac domains that showed associations with tau similar in direction and strength. These segments reflected type A/B compartment structure derived from Hi-C data as indicated by the red and blue ribbon at the bottom. The lower ribbon indicates lamina-associated domains (LADs) which are enriched in type B compartments.
|
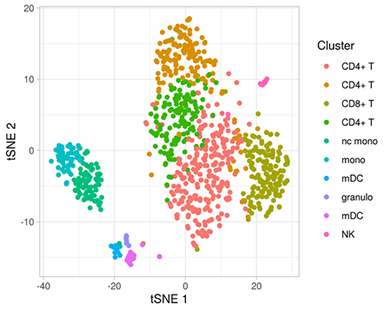
Figure 2: Clustering of 902 CSF cells from a person with mild cognitive impairment. Based in their transcription profiles (6328 detected genes), cells were clustered into 10 cell types as indicated by different colors and plotted in a low-dimensional representation.
Interestingly, utilizing tau over-expressing induced pluripotent stem cell-derived neurons, we demonstrated that tau-related chromatin alterations could be attenuated by a small molecule computationally predicted to reverse the tau effect. In collaboration with Zhiguo Zhang (Institute for Cancer Genetics), we will characterize in more detail the molecular mechanisms whereby tau impacts the nucleosome assembly of H3.3 and heterochromatin in AD neurons.
Other ongoing collaborative studies include a pilot project funded by the ADRC: Drs. Lawrence Honig, Marta Olah and I are generating a high-resolution multi-omic single-cell atlas of the cerebrospinal fluid (CSF) immunity in AD (Figure 2). One question we want to address with this dataset is whether the inflammatory expression signature that we previously observed in microglia cells from AD brains can also be detected in specific CSF cells. In a separate project with fellow faculty members Drs. Philip De Jager and Martin Picard, we utilize multi-omic brain data from the ROSMAP study of aging and dementia to characterize mitochondrial DNA copy number and heteroplasmy in AD brains and their relation to pathologies and molecular endophenotypes.
Hans-Ulrich Klein, PhD
Assistant Professor of Neurological Sciences (in Neurology, the Taub Institute for Research on Alzheimer's Disease and the Aging Brain)
Center for Translational and Computational Neuroimmunology (CTCN)
hk2948@cumc.columbia.edu